Central Theme: The role of epigenetics in fertility and infertility - from gametes to implantation and placentation
Introduction
Welcome to the UCSF Center for Research, Innovation and Training in Reproduction and Infertility!
We are one of eight Centers in the NIH Eunice Kennedy Shriver National Institute for Child Health and Human Development (NICHD) P50 National Centers in Translational Research in Reproduction and Infertility (NCTRI). These centers form a national network of multidisciplinary teams that promote bidirectional knowledge transfer between the laboratory and clinical translation with the ultimate goal to improve human reproductive health through research excellence, discovery, innovation and training.
The Center for Reproductive Sciences has awarded funding for three Pilot Project Proposals from the UCSF NIH National Center for Translational Research in Reproduction and Infertility (NCTRI). These Pilot projects are related to the Center’s central theme of early human development and the origins and biological consequences of human infertility. Paolo Rinaudo, Carol Anderson, and Nadia Roan were recipients of the awards for this year.
Background
Infertility affects about 7% of married couples in the U.S. It is recognized as a disease by the World Health Organization and causes significant morbidity and compromised quality of life among those affected. Despite the wide-spread prevalence of infertility of which about 1/3 is attributable to female factors, 1/3 male, and 1/3 both, mechanisms underlying the causes of infertility and translating this knowledge into personalized therapies and accurate diagnostics are wanting. Treatments are mostly empiric because mechanisms resulting in infertility, e.g., ovarian aging, oligo/anovulation, gonadal failure, poor oocyte quality, abnormal sperm, implantation failure, recurrent miscarriage, endometriosis, abnormal genital tract anatomy or hormonal response and inflammation, are rarely known with certainty. Also, pregnancies in women with infertility, either naturally conceived or result from infertility treatments, have greater risk of complications due to the underlying infertility or infertility therapies and include pre-eclampsia, preterm birth, and fetal growth restriction. These can have life-long effects on the health of children and adults born from these therapies and applied technologies. Thus, understanding mechanisms underlying reproductive success and compromise at the genomic, molecular, and cellular levels is crucial to fertility and the health and well-being of this and future generations. Also, engaging investigators from multiple disciplines is essential to unravel the complexities of successful reproduction. Equally important is building a sustainable pipeline of junior investigators in this field and promoting public literacy about reproductive health and science. These are core principles of our UCSF NCTRI Center, funded 2007-2023, directed by PI Linda Giudice, MD, PhD, Distinguished Professor of Obstetrics, Gynecology and Reproductive Sciences, and Associate Director, Marco Conti, MD, Professor of Obstetrics, Gynecology and Reproductive Sciences and immediate past Director of the UCSF Center for Reproductive Sciences.
The overall goals of our UCSF Center for Research, Innovation and Training in Reproduction and Infertility are:
- To advance research in reproductive science and medicine through trans-disciplinary collaboration and scientific and technologic innovation to improve human reproductive health and fertility.
- To serve as a national resource to inspire and mentor early career investigators in reproduction and infertility research and nurture their career development long-term.
- To communicate and be a national resource for reproductive research and relevance to reproductive health and fertility for the public, health care professionals, and patients.
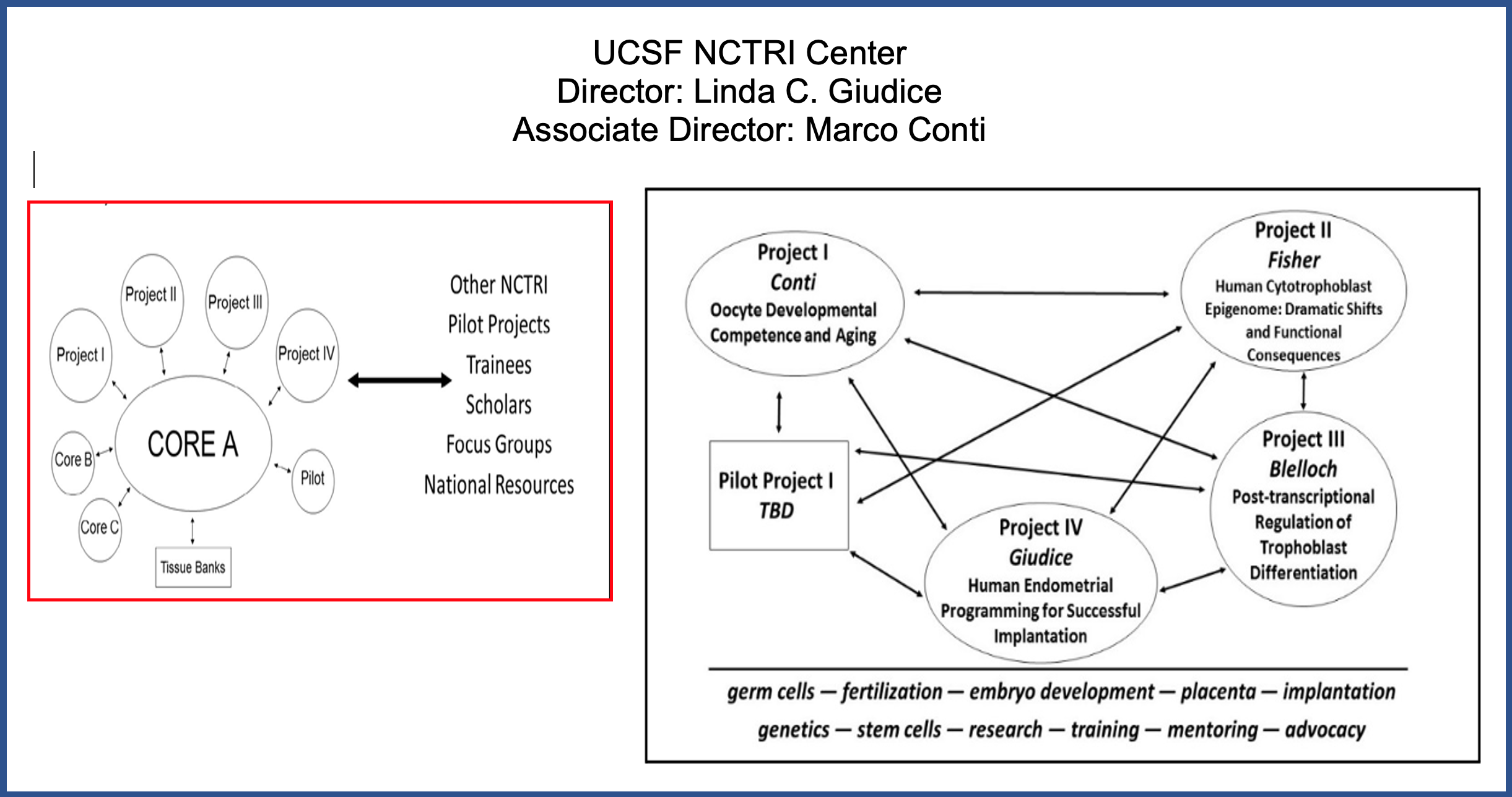
To accomplish these aims, our UCSF NCTRI Center unites our transdisciplinary expertise in basic and translational investigation in reproduction with our longstanding commitment to professional training and innovation. The central theme of our Center is the role of epigenetics in fertility and infertility - from gametes to implantation and placentation. Our Center is comprised of 4 research projects and pilot projects, supported by an administrative core, a multidimensional quantitative imaging core, an education, community outreach core, and a human uterine research biorepository bank.
Our multidisciplinary team of senior and junior researchers and trainees investigates molecular, genetic, and epigenetic mechanisms involved in acquisition of oocyte competence (Project 1, Conti), placental trophoblast biology (Project 2 Fisher, Project 3, Blelloch), and endometrial receptivity and paracrine interactions between the trophoblast and maternal decidua (Project 4, Giudice). We collaborate with colleagues with complementary expertise in immunology, genomics, epigenomics, and informatics. We have a strong track record of training students and fellows in our labs, and through our education, community outreach core C (Mellon), we continue mentorship and bringing reproductive science to our community. Our projects use advanced technologies and “omics” approaches, single cell analyses, animal models and human tissues and cells, and integrate well-annotated, relevant human phenotypic and clinical data to inform our studies. Our immediate goals are to determine epigenetic regulation of processes resulting in successful reproduction and infertility. Our long-term goals are to develop diagnostics and targeted therapeutics to alleviate infertility and poor reproductive outcomes and enhance the well-being of those with infertility and reproductive compromise.
Our UCSF NCTRI administrative Core resides in the administrative offices of the UCSF Center for Reproductive Sciences (CRS), Interim Director, Sindy Mellon, PhD. Our faculty and trainees participate in the annual CRS Reproductive Research day and bi-weekly CRS workshops, and trainees have full advantage of all UCSF in person and virtual seminars and webinars, educational opportunities, and mentoring and training opportunities.
NCTRI Faculty
 |
Project 1: Oocyte Developmental Competence and Aging Co-Investigators: Laird, Rinaudo; Collaborators: Blelloch, Fisher, Giudice |
The overall aim of the research projects in Dr. Conti’s laboratory is to understand the molecular mechanisms that control the development of the female gamete. In the past, the laboratory has contributed to the identification of the signaling pathways that control oocyte meiotic arrest and reentry into the meiotic cell cycle. Several key components of the cell cycle were identified and characterized by Dr. Conti’s team. More recently, the laboratory has focused on understanding how gene expression is regulated in the oocyte during growth, during maturation, and at the transition to the totipotent cell, the zygote. Synthesis of mRNA becomes virtually undetectable in fully grown oocytes arrested in meiotic prophase. Thus, gene expression during these final stages of oocyte maturation relies on an elaborate program of storage of maternal mRNAs synthesized early during oocyte growth, their recruitment to translating ribosomes, and eventually their degradation. The Conti lab was the first to provide a genome-wide insight into this program of translation and repression of maternal mRNAs and identified novel functions for known RNA binding proteins. More recently, we investigated the temporal dimension of maternal mRNA recruitment to the translation machinery which has led to the discovery of a switch in the mRNA translation program in the oocyte. Translation of mRNAs involved in oocyte growth becomes repressed and is replaced by activation of translation of mRNA coding for cell cycle components and for the transcription and chromatin remodeling machinery that will be used during embryo development. Our use of several different experimental models has established that defects in the execution of this switch in the translation program invariably compromise the oocyte’s fitness to support fertilization and embryo development. Recent findings also show that the decline in oocyte quality with maternal aging is associated with defects in the program of translation of maternal mRNAs, a research directive actively pursued in the lab. Together with this project on maternal aging, the Conti lab is focusing on the molecular components involved in the activation and repression of translation of maternal mRNAs, as well as the role of polyadenylation in directing their destabilization. An additional project focuses on heterogeneity of the 3’UTR of mRNAs accumulated in the oocyte during development and the effect of these different 3’UTRs on the translation of encoded proteins.
|
Project 2: The Human Cytotrophoblast Epigenome: Dramatic Shifts and Functional Consequences Co-investigators: Drs. Blelloch, Costello, Giudice and Erlebacher |
Susan Fisher, PhD, is Professor in the Department of Obstetrics, Gynecology and Reproductive Sciences. She has over 35 years of experience studying the human placenta in the context of normal pregnancy and the major pregnancy complications. She and her group have developed in vitro models that have allowed them to study the role of proteinases, cell-matrix interactions, angiogenic/vasculogenic regulators, Eph/ephrin family members, Notch signaling and immune molecules in normal placentation. The results of these studies enabled parallel analyses of abnormal placentation in preeclampsia (PE) and preterm birth (PTB}, the common trisomies (13, 18 and 21) and placenta accreta spectrum disorder. Additionally, they have been using global transcriptional profiling to identify changes in placental/trophoblast gene expression as a function of gestational age and as a consequence of the aforementioned pregnancy complications. Companion global protein expression data (generated using mass spectrometry approaches, see below) has enabled prioritization of potential differentiation or disease regulators for functional analyses. In the last several years, they extended these analyses to an epigenetic level, collaboratoring with Dr. Joseph Costello, incorporating bisulfite-seq, ChIP-seq and ATAC-seq technologies into the methods they routinely apply. In parallel, they expanded their capabilities to include profiling specific subpopulations of cells at the maternal-fetal interface that are captured by laser microdissection. This approach revealed a great deal of novel information about the gene and protein expression patterns of these cell types that was obscured when the placenta/basal plate as a whole was profiled as the signals were diluted by other cell types, which are more numerous. Recently, she and her group have worked with Dr. Robert Blelloch to develop methods for applying single cell RNA-seq approaches to the human placenta and smooth chorion. With Dr. Linda Giudice they showed that cytotrophoblast-derived extracellular vesicles have interesting effects on human decidual cells that are mediated, in part, by TNFa. With Dr. Adrian Erlebacher, they have been studying mechanisms of maternal-fetal (placental) tolerance.
Dr. Fisher has dual expertise in mass spectrometry (MS)-based analyses, having completed a 5-year postdoctoral fellowship in this area. She has over four decades of experience in biological MS. Throughout her career, a major focus has been glycan analyses, specifically the structural characterization of human complex oligosaccharides. Her group is also very experienced in MS-based methods of protein analysis (glycosite mapping, sequential window acquisition of all theoretical fragment ion spectra [SWATH]).
 |
Project 3: Post-transcriptional Regulation of Trophoblast Differentiation Co-Investigators: Fisher, Sirota/Fung; Collaborators: Conti, Giudice |
Dr. Robert Blelloch received his MD and PhD degrees from the University of Wisconsin-Madison. While studying for his PhD under the mentorship of Judith Kimble, PhD he discovered a novel matrix metalloproteinase, which is essential for normal gonad morphogenesis in the C. elegans nematode. After earning his dual degree, Blelloch completed a residency in clinical pathology followed by a fellowship in transfusion medicine at Brigham and Women’s Hospital. During his residency and fellowship Blelloch concurrently conducted research at the Whitehead Institute of Biomedical Research under the mentorship of Rudolf Jaenisch, MD. Through this research, Blelloch developed an interest in the epigenetic regulation of normal development and disease. In particular, he used somatic cell nuclear transfer (SCNT) to globally evaluate the role of epigenetics in cancer. This approach enabled him to show a critical, although variable, role for epigenetics in tumor development and maintenance. He also used SCNT to show a role for global DNA methylation in the maintenance of cellular differentiation. Since starting his own lab in 2006, Blelloch continues to follow his interest in the global regulators of cellular differentiation and cancer. His work is focused on the role of small noncoding RNAs in stem cell regulation and prostate cancer. Blelloch's ultimate goal is to develop a deeper understanding of the molecular underpinnings of normal and abnormal cell differentiation which will contribute to the advancement of regenerative medicine and cancer care. Blelloch is a professor in three departments: Urology; Obstetrics, Gynecology and Reproductive Sciences; and Pathology. He holds the Peter R. Carroll, MD Endowed Chair in Urology. Blelloch is a member of the Eli and Edythe Broad Center of Regeneration Medicine and Stem Cell Research, the Center for Reproductive Sciences, and the UCSF Helen Diller Family Comprehensive Cancer Center. In 2013, Blelloch was named Associate Chair, Basic Science Research for the Department of Urology.
Research Interests
Our laboratory is interested in determining the molecular mechanisms that regulate stem cell differentiation and de-differentiation and how these mechanisms become deregulated in cancer. Since starting our laboratory in 2006, we have focused on the role of post-transcriptional regulators including microRNAs (miRNAs) and RNA binding proteins (RBPs). This work is important in embryonic stem cell development and in the development of other cell types, including prostate cancer cells. (See Research for more details.)
|
Project 4: Human Endometrial Programming for Successful Implantation Co-Investigators: Fisher, Fung, Sirota, Roan; Collaborators: Conti, Blelloch, Erlebacher, Rabban |
Dr. Giudice is Distinguished professor of OBGYN/RS and is a reproductive endodcrinologist and biochemist. Our laboratory studies human endometrium (the lining of the uterus), which is critical for pregnancy success and, as a mucosal tissue, is a sentinal against infection in the upper female reproductive tract. We pursue biochemical, molecular, and multi-omics approaches to investigate the biology of human endometrium and mechanisms underlying steroid hormone responses of its cellular components in health and disease (endometriosis, PCOS, uterine fibroids). We also study interactions between the trophoblast and endometrium relevant to infertility and pregnancy outcomes, endometrial and systemic myeloid populations in endometrial disorders, stem/progenitors in cyclic endometrial regeneration and tissue homeostasis, and effects of contraceptive steroids on the upper female reproductive tract relevant to HIV transmission risk and tissue dyshomeostasis. We use in vitro cultures and isolated cell and bulk tissue analyses, whole genome transcriptomic, epigenomic, and single cell technologies, and advanced bioinformatic approches to understand functions and dysfunctions of endometrial epithelial, endothelial, mesenchymal, and immune lineages, normally and in reproductive disorders. These have resulted discovery of unique transcriptomic signatures, mediators of cell-cell cross-talk, signaling pathways and upstream regulators that guide functional studies. In our P50 NCTRI Center, the Specific Aims of Prpoject 4 are to:
1) determine if genome-wide epigenetic programming of endometrial stromal fibroblasts (eSF) is inherited from its progenitor (mesenchymal stem cell (eMSC)) and responds to steroid hormones affecting eSF functionality; 2) Determine endometrial immune features in endometriosis and effects on eMSC differentiation and eSF decidualization; 3) test whether endometriosis results in abrogated eSF amplification of signals from placental cytotrophoblasts.
As a physician scientist, my lab’s focus is on translational science, and most of our research is targeted to this goal and training and mentoring the next generation of reproductive biologists and reproductive medicine investigators.
 |
Multidimensional Imaging Core |
Drs. Jennifer Fung and Dr. Marina Sirota provide the expertise for the Multidimensional Imaging Core. Dr. Fung has extensive experience in the development of several imaging techniques and as well microarray/sequencing technology for the study of meiosis. We are currently developing new assays to perform high-throughput reproductive toxicity assessments. Dr. Sirota has an in-depth background in bioinformatics and has worked closely with Dr. Giudice on many endometriosis related projects.
The multidimensional imaging core provides unique imaging and image analysis capabilities to investigators within our P50 NCTRI Center. Center investigators have free access to a INCell 6000 confocal imaging platform in which high-throughput fluorescence imaging can be performed. The imaging core develops software for automated image analysis for the INCell 6000 as well as custom image analyses for data obtained using other microscopy modalities. The core develops custom machine learning classification and segmentation algorithms for individual projects. In addition, the core provides statistical and pipeline development expertise for both RNA and DNA sequencing projects.
 |
Education/Outreach Core |
Synthia Mellon, Professor of Ob, Gyn, is Interim Director of the UCSF Center for Reproductive Sciences and Director of the Education/Outreach Core of our UCSF NCTRI Center. Our laboratory studies the regulation of steroid hormone synthesis, by analyzing the transcriptional regulation of the steroidogenic enzymes, and the mechanism of action of steroids in both steroidogenic tissues and in the nervous system, where we identified the developmental and regional expression of steroidogenic enzymes and some novel actions of steroids on neuronal function. Steroid hormones are regulators of a multitude of physiologic processes, and act primarily by activating nuclear receptors, which are transcriptional regulators of various genes. However, in the nervous system, where we found steroidogenic enzymes and synthesis of a novel class of steroid, called neurosteroids, which modulate ion flux through the ion gated GABAA and NMDA receptors, and regulation of their synthesis results in changes in behavior, learning, and memory. We delineated the ontogeny and sites of steroidogenic enzyme expression and showed that neurosteroids affect neuronal development by specifically modulating either axonal or dendritic growth and neuronal differentiation. Their actions throughout life may maintain the integrity of neural connections, and demise of neurosteroid synthesis may result in loss of memory associated with some age-related neurologic diseases. We expanded these studies to understanding the role of neurosteroids in human neuropsychiatric diseases (major depressive disorder, post-traumatic stress disorder), and are studying other biological consequences and causes of these disorders, especially mitochondrial dysfunction and neuroinflammation. Our studies thus rely on a number of different experimental paradigms, from molecular biologic analysis of gene structure and transcription factors to developmental and cell biology of neuronal development and function.
The goal of Core C in our UCSF NCTRI is to develop a structured educational outreach and mentoring program to promote research in reproductive sciences, increase scientific literacy, and engage the San Francisco Bay Area community. We have built upon well-established programs at UCSF: the Science and Educational Partnership (SEP); a program developed in our Department of OB/GYN/Reproductive Sciences (URI, Women’s Health Undergraduate Research Internship); the San Francisco State Bridges Program, and various other summer research programs at UCSF, to train young investigators in the labs of NCTRI scientists, while also providing an opportunity for graduate students and postdoctoral fellows to learn how to be effective mentors. We have expanded our outreach to include a new program established by one of our pilot PIs (PROPEL post-baccalaureate program, established by Todd Nystul) to reach underserved college graduates who need more hands-on laboratory training before applying to graduate programs. We have also participated annually in the Bay Area Science Festival, a widely attended science festival geared toward children and their families, promoting our research in reproduction, with a game-filled flare.
 |
Pilot Project: Single-Cell Analysis of the Altered Placental Epigenome in Sporadic Miscarriage |
I received training in both basic sciences and clinical research (pediatrics and neonatology). My research integrates multi-omics data to identify genetic underpinnings of complex diseases. My research introduced evolutionary insights into clinical research and illustrated the role of population differentiation underlying the stark population disparity of neonatal prematurity. My research identified genes in bronchopulmonary dysplasia (BPD, a severe lung disease prevalent in premature newborns). My study also uncovered the contribution to neonatal preterm birth from fetal de novo mutations, preferentially affecting genes in early brain development. My laboratory at UCSF is studying gene environmental interactions underlying health disparities of preterm birth among human populations. Extending our research interest from late gestation events to early gestation, as part of the P50 efforts, my group is now dedicated to integrating large-scale miscarriage patient genomes with single-cell placental epigenomes in the first trimester to fine-map the altered epigenomic regulatory elements and to identify the most vulnerable cell types in miscarriage. Our studies will provide valuable resources to the community and will help reveal molecular mechanisms of pregnancy complications at a single cell resolution.
Research Overview: Health is a descriptive concept, which is often defined as “free from disease”. Our research aims to build a data-driven framework to quantitatively define health. We develop machine learning algorithms to define personal genome baselines for disease occurrences, and identify actionable lifestyle elements whose modification could compensate for genetic risk. We integrate personal genome baselines and individual lifestyles for precision health management.
Large-scale analysis of disease genomes: Because many complex human diseases have a strong genetic component, it has been long anticipated that once we can sequence one’s personal genome, we would be able to predict the person’s clinical outcome. This concept has been the cornerstone of today’s precision medicine. However, our genomic analysis of complex diseases is often challenged by mutational heterogeneity, where different patients often carry different sets of clinical mutations. We recently showed that these seemingly heterogeneous mutations are in fact convergent onto common pathways. We therefore use multi-omic profiling strategies to construct biological networks, and map genomic mutations onto biological networks to identify disease-associated pathways.
Clinical phenotyping using emerging technologies: In addition to using clinical records to model gene-environment interaction, we also leverage imaging and wearable sensor technologies to longitudinally digitize clinical traits. We use these technologies to derive quantitative traits for fine-mapping of genetic elements. We also perform digital signal processing to analyze these longitudinal data to capture subtle signals that will enable us to detect diseases before symptoms emerge. With comprehensively profiled individual lifestyles and physiologies, we aim to extend the classical P=G+E model to a Bayesian framework, where we stratify individuals based on personal genome baselines, followed by identifying lifestyle elements whose modification could improve clinical outcome.
Using iPSC-derived cells to identify pathogenic non-coding mutations: Our current understanding of disease genomes is largely limited to coding sequences, and the non-coding regions, accounting for ~98.5% of the human genome, have been largely unexplored for their contribution to disease etiologies. We perform ChIP-Seq and ATAC-Seq in iPSC-derived cells to delineate the genome-wide chromatin architecture in disease-related cell types, and build machine learning algorithms to identify non-coding mutations (SVs, indels and CNVs) that disrupt local and global chromatin architecture.
Primary Thematic Area: Human Genetics
Secondary Thematic Area: Developmental & Stem Cell Biology
Research Summary: Large-scale analysis of disease genomes by integrating multi-omics data, evolutionary insights, health records, and digitized clinical traits from imaging and wearable sensor readouts.
  |
Pilot Project: |
Our laboratory uses the Drosophila ovary as a model for studying the mechanisms that govern cell fate during oogenesis and epithelial stem cell self-renewal.
We are interested in questions such as:
1. How is stem cell fate maintained within a dynamic epithelial tissue?
2. What is the nature of the epithelial stem cell niche?
3. What is the role of epithelial stem cells in normal reproductive physiology?
4. How do chemical and morphological features of cells contribute to cell fate?
These studies have led to new insights into topics in adult stem cell biology, development, and reproduction, such as the genetic regulation of stem cell niche competition, the role of pHi dynamics in cell fate, and the role of actin dynamics in the differentiation of embryonic stem cells.
The follicular epithelium in the Drosophila ovary is an ideal model for the study of epithelial biology. It possesses many classical epithelial features, such as a columnar cell shape, apical/basal polarity, and canonical cell adhesion complexes, and yet is a relatively simple tissue and is highly tractable for molecular and cell biological analysis. Combined with the powerful genetic tools available in Drosophila, this allows us to address questions in epithelial stem cell and tissue biology with single-cell resolution in the natural, in vivo context.
Major Goals: Our laboratory uses the Drosophila ovary as a model for studying the fundamental properties of epithelial stem cells, their associated niche, and the connection between epithelial stem cells and cancer. We are interested in questions such as:
1. How is stem cell fate maintained within a dynamic epithelial tissue?
2. What is the nature of the epithelial stem cell niche?
3. What is the role of epithelial stem cells in normal reproductive physiology?
4. Do stem cell defects underlie epithelial cancers and what can studies of epithelial stem cells teach us about the earliest steps in cancer formation?
Ongoing research:
Identifying Stem Cells and their Niche: Although adult stem cells are believed to reside in distinct microenvironments, or niches, that function to regulate stem cell behavior, niches have been hard to study because of the difficulty of precisely identifying the stem cells in most tissues. However, we have developed a set of criteria that facilitates reliable identification of the epithelial follicle stem cells (FSCs) in the Drosophila ovary and we are now mapping their interactions with neighboring cells to better understand the nature of the FSC niche. Surprisingly, we have found that the FSC niche appears much more dynamic than the few previously characterized niches. We are using lineage analysis to follow FSC behavior, track the patterns of FSC daughter cell migration and differentiation, and investigate relevant gene function.
Epithelial Stem Cell Genetics: The wnt/wingless, hedgehog, BMP and Notch signaling pathways are all important for FSC function and early epithelial development but little is known about where in the process these signals exert their effects or how they are coordinated to produce a functional, healthy epithelium. We are investigating the function of key signaling components at specific steps in early follicle formation to map the contributions of these pathways to FSC function and follicle formation. In addition, we are interested in the role that misregulation of signaling in the ovarian epithelial cells plays in ovarian cancer. We are now investigating the interaction between the wnt pathway and other putative ovarian cancer genes in follicle cells as well as screening for markers that identify pre-tumorous cells to better understand the early steps leading to hyperplasia. Through collaboration with our colleagues in the Center for Reproductive Sciences, we will be able to test whether gene interactions and markers that we identify in the fly ovary will also be present in mammalian model systems and human tissues. To identify additional genes that are required for proper FSC function, we screened through a collection of over 600 lines bearing lethal mutations. We have identified several mutants with a follicle stem cell phenotype including ones that accelerate the rate of stem cell loss; confer a “hyper-competitive” stem cell replacement phenotype; and/or cause over-proliferation, perhaps modeling a precancerous state.
Epithelial Stem Cells and Oogenesis Oogenesis is well conserved from flies to mammals and studies of the Drosophila ovary have provided valuable insight into the process of female reproduction. Our work on the characterization of the follicle stem cells and their associated niche provides an opportunity to use the fly ovary as a model of follicle formation. By studying the lineage just downstream of the FSCs, we found that FSCs produce “pre-follicle cells” that are developmental intermediates between the FSCs and the polarized epithelium. Follicle formation begins when a newly formed germline cyst moves past the FSC niche and contacts these pre-follicle cells. We found that a Delta signal from the germline activates Notch in some pre-follicle cells, inducing them to migrate along the anterior fact of the cyst toward the opposite niche. Other pre-follicle cells that do not receive the signal instead migrate away from the niche toward the posterior, directly into the polarized epithelium. We are now studying how these and other cellular events lead to the formation of a new follicle during normal oogenesis.
Diane Barber
Research Overview: Our research program aims to understand basic cell biology processes and how they are dysregulated in diseases. Within this context we are determining how protein behaviors and cell functions are regulated by intracellular pH (pHi) and actin filament dynamics. We bridge structural and cell biology to reveal the molecular mechanisms of cell behaviors, with a focus on behaviors relevant to epithelial plasticity, particularly cancer cell biology and stem cell differentiation. The high impact our pioneering work is highlighted by our publication h-index of 46.
Cancer cell biology: Cancer cells have a higher pHi than normal cells, which we and others confirmed is necessary for disease progression. Our previous review on pHi and cancer (Webb et al., Nature Rev Cancer, 2009) has been cited by > 1250 publications. The molecular mechanisms mediating pHi-regulated cell behaviors, however, remain understudied and largely unknown. Our work bridges protein electrostatics and structure with cell biology to reveal how pHi dynamics regulates cell behaviors through protonation of titrating amino acids as a post-translational modification to regulate protein structure and function (Schönichen et al., 2013 Ann Rev Biophys. 42:289). We revealed the design principles and functions of “pH sensors” described as endogenous proteins regulated within the cellular pH range, including guanine nucleotide exchange factors regulating cell polarity (Frantz et al., 2007 J Cell Biol. 179:403), cofilin controlling actin assemblies (Frantz et al., 2008 J Cell Biol. 183:865), b-catenin regulating tumorigenesis (White et al., 2018 J Cell Biol. 217:3965), and talin (Srivastava et al., 2008 Proc Natl Acad Sci. 105:14436) and the focal adhesion kinase FAK (Choi et al., 2013 J Cell Biol. 202:849) controlling cell-substrate adhesion.
Stem cell differentiation: In collaboration with the Todd Nystul lab, we are determining how pHi and actin filament dynamics regulate stem cell differentiation, which remains understudied. This is the basis of our UCSF NCTRI Pilot Project. Using multiple embryonic and adult stem cell models we showed that an increase in pHi is necessary for stem cell differentiation and also lineage specification, in part by regulating Wnt pathway activity (Ulmschneider et al., 2016 J Cell Biol. 215:345, Benitez et al., 2019 Dev Biol. 452:127, Yi et al., 2022 BIORXIV/2021/466337). A current objective is to identify pH sensors necessary for stem cell differentiation, aligned with our report on pHi dynamics and b-catenin stability (White et al., 2018 J Cell Biol. 217:3965) and our work on how pHi regulates transcription factor-DNA binding selectivity. We also are characterizing how remodeling of actin filaments is necessary for differentiation of naïve embryonic stem cells to the primed state with a focus on how actin dynamics is linked to transcriptional events.
A particular interest is interfacing our work on cancer and stem cell biology through the adage that cancer is development gone wrong.
 |
Pilot Project: Role of Seminal Plasma in Promoting Endometrial Decidualization in Health and Disease |
Nadia Roan, PhD, received her undergraduate training from the University of California Berkeley, where she graduated with an honors degree in Molecular and Cellular Biology. She then completed her PhD in the Biological and Biomedical Sciences Program at Harvard Medical School studying mucosal T cell responses to the bacterium Chlamydia trachomatis. This was followed by postdoctoral studies at the Gladstone Institutes, where she studied the role of semen factors in reproductive health and HIV transmission. Dr. Roan is currently Professor of Urology at UCSF and Senior Investigator at Gladstone Institutes, where her lab studies the interactions between immune cells and viruses, in the context of HIV transmission through mucosal tissues, HIV persistence and pathogenesis, and COVID-19 disease and immunity. Her lab is also actively engaged in studies understanding the role of T and NK cells in reproductive health and in the context of endometrial diseases associated with female infertility. For their studies, the Roan lab implements a variety of high-dimensional single-cell analysis and bioinformatics tools on primary cells obtained from patient specimens and ex vivo tissue models. Studies in her lab funded by the P50 pilot program have demonstrated that proteins associated with extracellular vesicles from seminal plasma promote decidualization of endometrial stromal fibroblasts in a manner dependent on IL11 signaling, and ongoing studies are using single-cell RNAseq to characterize the signaling pathways induced during this process.
 |
Pilot Project: |
Dr. Sirota is Associate Professor at the Bakar Computational Health Sciences Institute at UCSF. Prior to that she was the Lead Research Scientist in the Division of Systems Medicine at Stanford University and has worked as a Senior Research Scientist at Pfizer where she focused on developing Precision Medicine strategies in drug discovery. She completed her PhD in Biomedical Informatics at Stanford University, where her graduate work focused on predicting drug-disease relationships based on gene expression to identify novel therapeutic indications for known drugs. Her research interests lie in developing computational integrative methods and applying these approaches in the context of disease diagnostics and therapeutics. Her primary focus is on leveraging and integrating different types of omics and clinical data to better understand the role of the immune system in disease. The Sirota laboratory is funded by NIA, NLM, NIAMS, Pfizer, March of Dimes, and the Burroughs Wellcome Fund. As a young leader in the field, she has been awarded the AMIA Young Investigator Award in 2017. Dr. Sirota also is the director of the AI4ALL program at UCSF, with the goal of introducing high school girls to applications of AI and machine learning in biomedicine. She is PI and Director of the UCSF Center for Prematurity Research funded by the March of Dimes.
Our Pilot project in the UCSF NCTRI Center studies human endometrium in patients with implantation failure. Embryo implantation is a complex event that relies on the coordination between the embryo and the endometrium. The process of implantation is not thoroughly understood but is thought to be a gradual process where an embryo adheres to the luminal surface of the endometrium, followed by invasion of the embryonic trophectoderm cells into the deeper layer of the endometrium. Implantation potential is therefore dependent on both embryonic factors (genetic composition, expression of critical adhesion molecules, trophectoderm differentiation, and adequate invasion) and endometrial receptivity. An underlying endometrial cause is often suspected in scenarios where euploid, excellent-quality embryos repeatedly fail to implant and progress into a clinical pregnancy, a condition known as recurrent implantation failure (RIF). There is no consensus regarding the absolute definition of RIF. The most commonly used cut-offs for RIF are failure to achieve clinical pregnancy after transfer of a total of 3 to 4 good-quality embryos1. Recently, much effort has been made to assess different aspects of endometrial receptivity in hopes of optimizing subsequent embryo transfer outcomes for patients. One such aspect is the evaluation of B-cell CLL/lymphoma 6 (BCL6) in the human endometrium, which is a proposed biomarker of endometriosis that is highly overexpressed in women with endometriosis during the secretory phase of the menstrual cycle. BCL6 is also highly expressed in many women with unexplained infertility and failed IVF attempts3. Further, this finding may significantly impact clinical practice, as evidence suggests dramatic improvement in clinical pregnancy and live birth outcomes with medical (GnRH agonist) or surgical (laparoscopy) treatment prior to embryo transfer in women found to have BCL6 over-expression. Our study aims to evaluate the transcriptomic landscape of the human endometrium in women experiencing infertility to better understand if BCL6 over-expression correlates on a single cell level, which could help explain and identify additional biomarkers of suboptimal endometrial receptivity.
 |
Pilot Project: |
Dr. Paolo Rinaudo is a specialist in reproductive endocrinology and fertility at the UCSF Center for Reproductive Health. In addition to caring for patients with fertility and reproductive endocrine disorders, Dr. Rinaudo conducts research on pre-implantation embryo development and the long-term consequences of in vitro embryoculture. He is the recipient of numerous honors and awards, including the ASRM/Serono grant in reproductive biology and the President Presenter Award at the 50th annual meeting of the Society for Gynecological Investigation, Washington, DC. He is a member of professional societies, such as the American Society for Reproductive Medicine, American College of Obstetrics and Gynecology and Society for Reproductive Endocrinology and Infertility. Rinaudo earned a medical degree at Torino Medical School and a doctorate at Torino University in Torino, Italy. He completed a residency in obstetrics and gynecology at Yale University, followed by a fellowship in reproductive endocrinology and infertility at the Hospital of the University of Pennsylvania. He is Professor of Obstetrics, Gynecology and Reproductive Sciences at UCSF. The focus of his basic science research is to understand how stress during the pre-implantation period affects fetal and adult development. This has particular relevance in view of the widespread use of assisted reproductive techniques, such as in vitro fertilization (IVF): more than 8 million children or 1.5% of the population of the Western World have been conceived by these technologies. Fetal adaptations in utero to adverse conditions can lead to specific diseases in the adult, including diabetes, high blood pressure and coronary heart disease - a phenomenon termed developmental origins of adult health and disease or the Barker hypothesis. His laboratory has created a mouse model of IVF, wherein the phenotype of mice conceived in vivo or in vitro is analyzed and compared. We have found that embryo and placenta development are altered following conception by IVF. Further, IVF offspring display abnormalities of postnatal growth, fat deposition and alterations of glucose homeostasis and lower lactate levels. Dr Rinaudo’s pilot project in our P50 NCTRI Center is aimed at generating preliminary data to discover how embryonic metabolism can affect the epigenetic landscape of preimplantation mouse embryos.
Related Link:
National Centers for Translational Research in Reproduction and Infertility (NCTRI) Website